As part of my phyloinformatics course I've put together a simple browser to navigate through TreeBASE. The inspiration comes from genome browsers (e.g., the UCSC Genome Browser) where the genome is treated as a linear set of co-ordinates, and features of the genome are displayed as "tracks".
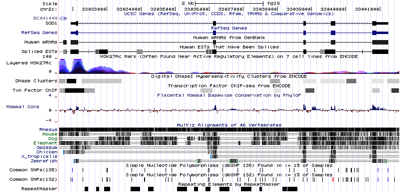
For my browser, I've used the order in which nodes appear in the NCBI tree as you go from left to right as the set of co-ordinates (actually, from top to bottom as my browser displays the co-ordinate axis vertically).
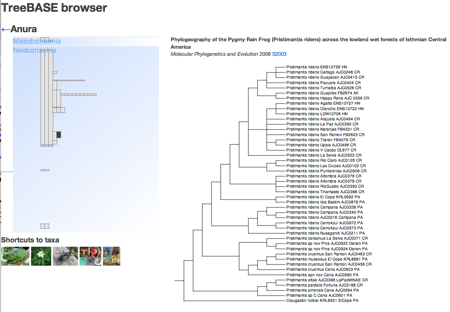
I then place each TreeBASE tree within this classification by taking the TreeBASE → NCBI mapping provided by TreeBASE and finding the "majority rule" taxon for each tree (in a sense, the taxa that summarises what the tree is about). Each tree is represented by a vertical line depicting the span of the corresponding NCBI taxon (corresponding to a "track" in a genome browser). Taking the majority-rule taxon rather than say, the span of the tree, makes it possible to pack the vertical lines tightly together so that they take up less space (the ordering from left to right is determined by the NCBI taxonomy).
If you mouse-over a vertical bar you can see the title of the study that published the tree. If you click on the vertical bar you'll see the tree displayed on the right (if your web browser understands SVG, that is). If you click on the background you will drill down a level in the NCBI classification. To go back up the classification, click on the arrow at the top left of the browser.
This is all very preliminary, but you can take it for a spin at http://iphylo.org/~rpage/phyloinformatics/treebase/.
Below is a short video walking you through some examples.